expan.core package¶
Submodules¶
expan.core.binning module¶
Binning module.
-
class
expan.core.binning.Binning¶ Bases:
objectThe Binning class has two subclasses: CategoricalBinning and NumericalBinning.
-
label(data, format_str=None)¶ This returns the bin labels associated with each data point in the series, essentially ‘applying’ the binning to data.
Parameters: - data (array-like) – array of datapoints to be binned
- format_str (str) – string defining the format of the label to apply
Returns: array of the bin label corresponding to each data point.
Return type: array-like
Note
Implemented in subclass.
-
-
class
expan.core.binning.CategoricalBinning(data=None, nbins=None)¶ Bases:
expan.core.binning.BinningA CategoricalBinning is essentially a list of lists of categories. Each bin within a Binning is an ordered list of categories.
CategoricalBinning constructor
Parameters: - data (array-like) – array of datapoints to be binned
- nbins (int) – number of bins
-
categories¶ Returns list of categories.
Returns: list of categories Return type: array-like
-
label(data, format_str='{standard}')¶ This returns the bin labels associated with each data point in the series, essentially ‘applying’ the binning to data.
Parameters: - data (array-like) – array of datapoints to be binned
- format_str (str) –
string defining the format of the label to apply
Options:
- {iter.uppercase}, {iter.lowercase}, {iter.integer}
- {set_notation} - all categories, comma-separated, surrounded by curly braces
- {standard} - a shortcut for: {set_notation}
Returns: array of the bin label corresponding to each data point
Return type: array-like
-
labels(format_str='{standard}')¶ Returns the labels of the bins defined by this binning.
Parameters: format_str (str) – string defining the format of the label to return
Options:
- {iter.uppercase}, {iter.lowercase}, {iter.integer}
- {set_notation} - all categories, comma-separated, surrounded by curly braces
- {standard} - a shortcut for: {set_notation}
Returns: labels of the bins defined by this binning Return type: array-like Note
This is not the same as label (which applies the bins to data and returns the labels of the data).
-
mid(data)¶ Returns the middle category of every bin.
Parameters: data – data on which the binning is to be applied Returns: the middle category of every bin Return type: array-like
-
class
expan.core.binning.NumericalBinning(data=None, nbins=None, uppers=None, lowers=None, up_closed=None, lo_closed=None)¶ Bases:
expan.core.binning.BinningThe Binning class for numerical variables.
- Todo:
Think of a good way of exposing the _apply() method, because with the returned indices, can then get uppers/lowers/mids/labels (ie reformat) without doing the apply again. Am experimenting with maintaining the lists with a single element tacked onto the end representing non-matching entries.
All access then are through properties which drop this end list, except when using the indices returned by _apply.
This means that the -1 indices just works, so using the indices to get labels, bounds, etc., is straightforward and fast because it is just integer-based array slicing.
NumericalBinning constructor.
Parameters: - data (array-like) – array of datapoints to be binned
- nbins (int) – number of bins
- uppers (array-like) – a list of upper bounds
- lowers (array-like) – a list of lower bounds
- up_closed (array-like) – a list of booleans indicating whether the upper bounds are closed
- lo_closed (array-like) – a list of booleans indicating whether the lower bounds are closed
-
label(data, format_str='{standard}')¶ Return the bin labels associated with each data point in the series, essentially ‘applying’ the binning to data.
Parameters: - data (array-like) – array of datapoints to be binned
- format_str (str) –
string defining the format of the label to apply
Options:
- {iter.uppercase} and {iter.lowercase} = labels the bins with letters
- {iter.integer} = labels the bins with integers
- {up} and {lo} = the bounds themselves (can specify precision: {up:.1f})
- {up_cond} and {lo_cond} = ‘<’, ‘<=’ etc.
- {up_bracket} and {lo_bracket} = ‘(‘, ‘[‘ etc.
- {mid} = the midpoint of the bin (can specify precision: {mid:.1f}
- {conditions} = {lo:.1f}{lo_cond}x{up_cond}{up:.1f}
- {set_notation} = {lo_bracket}{lo:.1f},{up:.1f}{up_bracket}
- {standard} = {conditions}
- {simple} = {lo:.1f}_{up:.1f}
- {simplei} = {lo:.0f}_{up:.0f} (same as simple but for integers)
- see:
- Binning.label.__doc__
- NumericalBinning._labels.__doc__
When format_str is None, the label is the midpoint of the bin. This may not be the most convenient. Might be better to make the default format_str ‘{standard}’ and then have the client use mid() directly if midpoints are desired.
-
labels(format_str='{standard}')¶ Returns the labels of the bins defined by this binning.
Returns: array of labels of the bins defined by this binning Options:
- {iter.uppercase} and {iter.lowercase} = labels the bins with letters
- {iter.integer} = labels the bins with integers
- {up} and {lo} = the bounds themselves (can specify precision: {up:.1f})
- {up_cond} and {lo_cond} = ‘<’, ‘<=’ etc.
- {up_bracket} and {lo_bracket} = ‘(‘, ‘[‘ etc.
- {mid} = the midpoint of the bin (can specify precision: {mid:.1f}
- {conditions} = {lo:.1f}{lo_cond}x{up_cond}{up:.1f}
- {set_notation} = {lo_bracket}{lo:.1f},{up:.1f}{up_bracket}
- {standard} = {conditions}
- {simple} = {lo:.1f}_{up:.1f}
- {simplei} = {lo:.0f}_{up:.0f} (same as simple but for integers)
Return type: array-like Note
This is not the same as label (which applies the bins to data and returns the labels of the data)
-
lo_closed¶ Return a list of booleans indicating whether the lower bounds are closed.
-
lower(data)¶ see upper()
-
lowers¶ Return a list of lower bounds.
-
mid(data)¶ Returns the midpoints of the bins associated with the data
Parameters: data (array-like) – array of datapoints to be binned Returns: array containing midpoints of bin corresponding to each data point. Return type: array-like Note
Currently doesn’t take into account whether bounds are closed or open.
-
up_closed¶ Return a list of booleans indicating whether the upper bounds are closed.
-
upper(data)¶ Returns the upper bounds of the bins associated with the data
Parameters: data (array-like) – array of datapoints to be binned Returns: array containing upper bound of bin corresponding to each data point. Return type: array-like
-
uppers¶ Return a list of upper bounds.
-
expan.core.binning.create_binning(x, nbins=8)¶ Determines bins for the input values - suitable for doing SubGroup Analyses.
Parameters: - x (array_like) – input array
- nbins (integer) – number of bins
Returns: binning object
-
expan.core.binning.dbg(lvl, msg)¶
-
expan.core.binning.isNaN(obj)¶
expan.core.debugging module¶
This started as a very simple central place to provide a really easy interface to output (standardised across projects) so I could always just do dbg(1, ‘blah’).
It has morphed and mutated into an ugly wrapper on top of logger.
I keep it because I still find logger to be a PITA and too complicated to use quickly.
- A few things it offers over just using logging:
- defaults to DEBUG level
- use positive and negative numbers to configure level
- standardised logging msg
- easier calling dbg(1, ‘blah’) - dubious advantage
Created on Aug 11, 2014
- Todo:
- Need to figure out how to tell the logging class to skip this module’s functions in the stack, for printing of module and function name etc.. Could potentially fiddle with the stack before calling logging, but that’s very dodgy.
@author: rmuil
-
class
expan.core.debugging.Dbg(logger=None, dbg_lvl=2)¶ Bases:
object-
get_lvl()¶ Returns debug level.
Returns: debug level
-
out(lvl, msg, skip_stack_lvls=0)¶ Note
This isn’t very well implemented for important messages... We should move to relying much more on the Logging class for dispatching and filtering logs.
Treat this as just a convenience.
Parameters: - lvl –
- msg –
- skip_stack_lvls –
-
reset()¶ Resets debug level and call count.
-
set_lvl(new_dbg_lvl)¶ Sets new debug level.
Parameters: new_dbg_lvl –
-
expan.core.experiment module¶
Experiment module.
-
class
expan.core.experiment.Experiment(baseline_variant, metrics_or_kpis, metadata={}, features='default', dbg=None)¶ Bases:
expan.core.experimentdata.ExperimentDataClass which adds the analysis functions to experimental data.
-
baseline_variant¶ Returns the baseline variant.
Returns: baseline variant Return type: string
-
delta(kpi_subset=None, derived_kpis=None, variant_subset=None, assume_normal=True, percentiles=[2.5, 97.5], min_observations=20, nruns=10000, relative=False, weighted_kpis=None)¶ Compute delta (with confidence bounds) on all applicable kpis, and returns in the standard Results format.
Does this for all non-baseline variants.
TODO: Extend this function to metrics again with type-checking
Parameters: - kpi_subset (list) – kpis for which to perfom delta. If set to None all kpis are used.
- derived_kpis (list) –
definition of additional KPIs derived from the primary ones, e.g. [{‘name’:’return_rate’, ‘formula’:’returned/ordered’}]
the original kpi column names are alphanumerical starting with a letter, i.e. [a-zA-Z][0-9a-zA-Z_]+ - variant_subset (list) – Variants to use compare against baseline. If set to None all variants are used.
- assume_normal (boolean) – specifies whether normal distribution assumptions can be made
- percentiles (list) – list of percentile values to compute
- min_observations (integer) – minimum observations necessary. If less observations are given, then NaN is returned
- nruns (integer) – number of bootstrap runs to perform if assume normal is set to False.
- relative (boolean) – If relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
- weighted_kpis (list) – a list of metric names. For each metric in the list, the weighted mean and confidence intervals are calculated, which is equivalent to the overall metric. Otherwise the metrics are unweighted, this weighted approach is only relevant for ratios.
Returns: Results object containing the computed deltas.
-
feature_check(feature_subset=None, variant_subset=None, threshold=0.05, percentiles=[2.5, 97.5], assume_normal=True, min_observations=20, nruns=10000, relative=False)¶ Compute feature check on all features, and return dataframe with column telling if feature check passed.
Parameters: - feature_subset (list) – Features for which to perfom delta. If set to None all metrics are used.
- variant_subset (list) – Variants to use compare against baseline. If set to None all variants are used.
- threshold (float) – p-value used for dismissing null hypothesis (i.e. no difference between features for variant and baseline).
- assume_normal (boolean) – specifies whether normal distribution assumptions can be made
- min_observations (integer) – minimum observations necessary. If less observations are given, then NaN is returned
- nruns (integer) – number of bootstrap runs to perform if assume normal is set to False.
Returns: pd.DataFrame containing boolean column named ‘ok’ stating if feature chek was ok for the feature and variant combination specified in the corresponding columns.
-
sga(feature_subset=None, kpi_subset=None, variant_subset=None, n_bins=4, binning=None, assume_normal=True, percentiles=[2.5, 97.5], min_observations=20, nruns=10000, relative=False, **kwargs)¶ Compute subgroup delta (with confidence bounds) on all applicable metrics, and returns in the standard Results format.
Does this for all non-baseline variants.
Parameters: - feature_subset (list) – Features which are binned for which to perfom delta computations. If set to None all features are used.
- kpi_subset (list) – KPIs for which to perfom delta computations. If set to None all features are used.
- variant_subset (list) – Variants to use compare against baseline. If set to None all variants are used.
- n_bins (integer) – number of bins to create if binning is None
- binning (list of bins) – preset (if None then binning is created)
- assume_normal (boolean) – specifies whether normal distribution assumptions can be made
- percentiles (list) – list of percentile values to compute
- min_observations (integer) – minimum observations necessary. If less observations are given, then NaN is returned
- nruns (integer) – number of bootstrap runs to perform if assume normal is set to False.
- relative (boolean) – If relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: Results object containing the computed deltas.
-
trend(kpi_subset=None, variant_subset=None, time_step=1, cumulative=True, assume_normal=True, percentiles=[2.5, 97.5], min_observations=20, nruns=10000, relative=False, **kwargs)¶ Compute time delta (with confidence bounds) on all applicable metrics, and returns in the standard Results format.
Does this for all non-baseline variants.
Parameters: - kpi_subset (list) – KPIs for which to perfom delta computations. If set to None all features are used.
- variant_subset (list) – Variants to use compare against baseline. If set to None all variants are used.
- time_step (integer) – time increment over which to aggregate data.
- cumulative (boolean) – Trend is calculated using data from start till the current bin or the current bin only
- assume_normal (boolean) – specifies whether normal distribution assumptions can be made
- percentiles (list) – list of percentile values to compute
- min_observations (integer) – minimum observations necessary. If less observations are given, then NaN is returned
- nruns (integer) – number of bootstrap runs to perform if assume normal is set to False.
- relative (boolean) – If relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: Results object containing the computed deltas.
-
-
expan.core.experiment.subgroup_deltas(df, variants, n_bins=4, binning=None, assume_normal=True, percentiles=[2.5, 97.5], min_observations=20, nruns=10000, relative=False)¶ Calculates the feature dependent delta.
Parameters: - df (pandas DataFrame) – 3 columns. The order of the columns is expected to be variant, feature, kpi.
- variants (list of 2) – 2 entries, first entry is the treatment variant, second entry specifies the baseline variant
- n_bins (integer) – number of bins to create if binning is None
- binning (list of bins) – preset (if None then binning is created)
- assume_normal (boolean) – specifies whether normal distribution assumptions can be made
- percentiles (list) – list of percentile values to compute
- min_observations (integer) – minimum number of observations necessary. If less observations are given, then NaN is returned.
- nruns (integer) – number of bootstrap runs to perform if assume normal is set to False.
- relative (boolean) – If relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: bin-name, mean, percentile and corresponding values list: binning used
Return type: pandas.DataFrame
-
expan.core.experiment.time_dependent_deltas(df, variants, time_step=1, cumulative=False, assume_normal=True, percentiles=[2.5, 97.5], min_observations=20, nruns=10000, relative=False)¶ Calculates the time dependent delta.
Parameters: - df (pandas DataFrame) – 3 columns. The order of the columns is expected to be variant, time, kpi.
- variants (list of 2) – 2 entries, first entry is the treatment variant, second entry specifies the baseline variant
- time_step (integer) – time_step used for analysis.
- percentiles (list) – list of percentile values to compute
- min_observations (integer) – minimum number of observations necessary. If less observations are given, then NaN is returned
- nruns (integer) – number of bootstrap runs to perform if assume normal is set to False.
- relative (boolean) – If relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: bin-name, mean, percentile and corresponding values list: binning used
Return type: pandas.DataFrame
expan.core.experimentdata module¶
ExperimentData module allows us to abstract the data format from the library such that we can have multiple fetcher modules that only need import this module, and none of our analysis modules need know anything about the fetchers.
In other words, this is the interface between data fetching and data analysis.
-
class
expan.core.experimentdata.ExperimentData(metrics=None, metadata={}, features='default', deepcopy=False)¶ Bases:
objectMain class in ExperimentData module.
-
mandatory_metadata¶ metadata that needs to be provided (default: {‘experiment’, ‘source’})
-
primary_indices¶ primary indices on which the analyses will be performed (default: [‘entity’, ‘variant’]
-
optional_kpi_indices¶ optional indices for analysis (default: [‘time_since_treatment’]
-
known_feature_metrics¶ metric names that are automatically considered as features
Want to be able to create results from just a single dataframe.
Parameters: - metrics – data frame that contains either KPI or feature
- metadata – the metadata dict
- features – either ‘default’, which searches the metrics data frame for predefined feature names or list, which subsets the metrics data frame with the given column indices or data frame, which is feature data frame itself and metrics is either KPI or None or None
- deepcopy – the internal data frames are, by default, shallow copies of the input dataframes: this means the actual data arrays underlying the frames are references to the input. In most use-cases, this is desired (reindexing will not reindex the original etc.) but it may have some edge-case issues.
-
__getitem__(key)¶ Allows indexing the ExperimentData directly as though it were a DataFrame composed of KPIs and Features.
-
feature_boxplot(feature, kpi, **kwargs)¶ Plot feature boxplot.
Parameters: - feature (list) – list of features
- kpi (list) – list of KPIs
- **kwargs – additional boxplot arguments (see pandas.DataFrame.boxplot())
-
feature_names¶ Returns list of features.
Returns: list of feature names Return type: set
-
filter_outliers(rules, drop_thresh=True)¶ Method that applies outlier filtering rules on an ExperimentData object inplace.
Parameters: - rules (dict list) – list of dictionaries that define filtering rules
- drop_thresh (boolean) – whether to remove added threshold columns (defaults to true)
Examples
First example shows a ‘rules’ example usage for the ‘threshold’ filter with two different kinds, ‘upper’ and ‘lower’ - anything lower than -10.0 is filtered by the first rule and anything higher than 10.0 is filtered by the second one.
>>> [ { "metric":"normal_shifted_by_feature", "type":"threshold", "value": -10.0, "kind": "lower" }, { "metric": "normal_shifted_by_feature", "type": "threshold", "value": 10.0, "kind": "upper" } ]
Second example shows the usage of additional ‘time_interval’ and ‘treatment_stop_time’ parameters (it implies that a ‘treatment_start_time’ column exists).
Given these parameters a per entity threshold is calculated by the following equation:
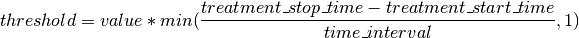
Using the equation above, the threshold defined for a specific time_interval is scaled according to the treatment exposure time for each entity. Thereby, the minimum scaling factor is 1, i.e. the time_interval defined in the outlier rules is the smallest threshold that is applied.
>>> [ { "metric": "normal_shifted_by_feature", "type": "threshold", "value": 1.0, "kind": "lower", "time_interval": 30758400, "treatment_stop_time": 30758500 } ]
Note
The outcome of the filtering depends on the order of rules being processed, e.g. when the rule contains a percentile instead of an absolute threshold.
- Todo:
- Implement other types of filtering, eg. percentile_XYZ_distribution.
-
known_feature_metrics= {'treatment_start_time', 'customer_age', 'exposed_customer', 'feature', 'gender', 'existing_customer', 'clv', 'orders_prev_year', 'orders_existing', 'zalando_age', 'sex', 'customer_zalando_age', 'start_segment', 'age'}
-
kpi_names¶ Returns list of KPIs.
Returns: list of KPI names Return type: set
-
mandatory_metadata= {'source', 'experiment'}
-
metric_names¶ Returns list of metrics (KPIs and features).
Returns: list of metric names Return type: set
-
metrics¶ Simply joins the KPIs and the features.
- Todo:
- It may well be worth investigating caching this result because the features and kpis will rarely change, and accessing them in this way is likely to be common.
-
optional_kpi_indices= ['time_since_treatment']
-
primary_indices= ['entity', 'variant']
-
-
expan.core.experimentdata.detect_features(metrics)¶ Automatically detect which of the metrics are features.
Parameters: metrics (pandas.DataFrame) – ExperimentData metrics Returns: dictionary of features present in the metrics Return type: dict
expan.core.results module¶
-
class
expan.core.results.Results(df, metadata={}, dbg=None)¶ Bases:
objectA Results instance represents the results of a series of analyses such as SGA or deltaKPI.
Q: could we make this a subclass of DataFrame (i.e. provide this class in an ‘is-a’ relationship with DataFrame, rather than the ‘has-a’ relationship it has now? It seems to be a difficult thing to subclass the DataFrame object for some reason. https://github.com/pydata/pandas/pull/4271 For now, will leave as a ‘has-a’ class.
- Todo:
- Can we remove the ‘value’ level from the columns, so that the columns of the dataframe are simply the names of the variants? This will make the columns a normal index rather than a multi-index. Currently, always a multi-index with second level only containing a single value ‘value’.
Want to be able to create results from just a single dataframe.
Parameters: - df (pandas.DataFrame) – input dataframe
- metadata (dict) – input metadata
- dbg –
-
append_delta(metric, variant, mu, pctiles, samplesize_variant, samplesize_baseline, subgroup_metric='-', subgroup=None)¶ Appends the results of a delta.
Modifies (or creates) the results data (df).
Parameters: - metric –
- variant –
- mu –
- pctiles –
- samplesize_variant –
- samplesize_baseline –
- subgroup_metric –
- subgroup –
-
binning¶ Return the binning object.
-
bounds(metric=None, subgroup_metric='-')¶ Parameters: - metric –
- subgroup_metric –
Returns:
-
calculate_prob_uplift_over_zero()¶
-
delta_means(metric=None, subgroup_metric='-')¶ Parameters: - metric –
- subgroup_metric –
Returns:
-
index_values(level='metric')¶ Return the metrics represented in this Results object
-
mandatory_column_levels= ['variant']¶
-
mandatory_index_levels= ['metric', 'subgroup_metric', 'subgroup', 'statistic', 'pctile']¶
-
relative_uplift(analysis_type, metric=None, subgroup_metric='-')¶ Calculate the relative uplift for the given metrics and subgroup metrics.
-
sample_sizes(analysis_type='delta', metric=None, subgroup_metric='-')¶ Parameters: - analysis_type –
- metric –
- subgroup_metric –
Returns:
-
set_binning(binning)¶ Store a binning object in the metadata.
-
sga_means(metric=None, subgroup_metric='-')¶ Parameters: - metric –
- subgroup_metric –
Returns:
-
sga_uplifts(metric=None, subgroup_metric='-')¶ Parameters: - metric –
- subgroup_metric –
Returns:
-
statistic(analysis_type, statistic=None, metric=None, subgroup_metric='-', time_since_treatment='-', include_pctiles=True)¶ This is just a basic ‘formatter’ to allow easy access to results without knowing the ordering of the index, etc. and to have sensible defaults. All of this can be accomplished with fancy indexing on the dataframe directly, but this should just serve as a convenience and an obvious place to ‘document’ what the typical use-case is.
For all arguments, None means all, and ‘-‘ means only those for which this particular argument is undefined (e.g. subgroup_metric=’-‘)
Parameters: - analysis_type (string) –
the type of analysis that produced the TODO: implement this! results (several can be present in a single Result object). Must be one of the following:
- ‘delta’: only those with no time_since_treatment information, and no subgroup defined
- ‘sga’: only those with subgroup defined
- ‘trend’: only those with time_since_treatment defined
- None: no restriction done
- statistic (string) – the type of data you want, such as ‘uplift’
- metric (string) – which metrics you are interested in
- time_since_treatment (int?) – TODO: implement
- include_pctiles (bool) – some statistics (e.g. ‘uplift’) can be present with percentiles defined, as opposed to just a mean. If this is true, they’ll be returned also. TODO: implement this!
- analysis_type (string) –
-
to_csv(fpath)¶ Persist to a csv file, losing metadata.
Parameters: fpath – file path where the csv should be created Returns: csv file Note
This will lose all metadata.
-
to_hdf(fpath)¶ Persist to an HDF5 file, preserving metadata.
MetaData is stored as attributes on a Group called ‘metadata’. This group doesn’t include any datasets, but was used to avoid interfering with the attributes that pandas stores on the ‘data’ Group.
- Args:
- fpath:
Returns:
-
to_json(fpath=None)¶ Produces either a JSON string (if there is no filepath specified) or a JSON file containing the results.
Parameters: fpath – filepath where the result JSON file should be stored Returns: JSON string with the results file: JSON file with the results Return type: string
-
uplifts(metric=None, subgroup_metric='-')¶ Parameters: - metric –
- subgroup_metric –
Returns:
-
variants()¶ Return the variants represented in this object
-
expan.core.results.delta_to_dataframe(metric, variant, mu, pctiles, samplesize_variant, samplesize_baseline, subgroup_metric='-', subgroup=None)¶ Defines the Results data frame structure.
Parameters: - metric –
- variant –
- mu –
- pctiles –
- samplesize_variant –
- samplesize_baseline –
- subgroup_metric –
- subgroup –
Returns:
-
expan.core.results.delta_to_dataframe_all_variants(metric, mu, pctiles, samplesize_variant, samplesize_baseline, mu_variant, mu_baseline, subgroup_metric='-', subgroup=None)¶ Defines the Results data frame structure.
Parameters: - metric –
- mu –
- pctiles –
- samplesize_variant –
- samplesize_baseline –
- mu_variant –
- mu_baseline –
- subgroup_metric –
- subgroup –
Returns:
-
expan.core.results.feature_check_to_dataframe(metric, samplesize_variant, mu=None, pctiles=None, pval=None, mu_variant=None)¶ Defines the Results data frame structure.
Parameters: - metric –
- samplesize_variant –
- mu –
- pctiles –
- pval –
- mu_variant –
Returns:
-
expan.core.results.from_hdf(fpath, dbg=None)¶ Restores a Results object from HDF5 as created by the to_hdf method.
Parameters: - fpath –
- dbg –
Returns:
-
expan.core.results.prob_uplift_over_zero_single_metric(result_df, baseline_variant)¶ Calculate the probability of uplift>0 for a single metric.
Parameters: - result_df (DataFrame) – result data frame of a single metric/subgroup
- baseline_variant (str) – name of the baseline variant
Returns: result data frame with one additional statistic ‘prob_uplift_over_0’
Return type: DataFrame
expan.core.statistics module¶
-
expan.core.statistics.alpha_to_percentiles(alpha)¶ Transforms alpha value to corresponding percentile.
Parameters: alpha (float) – alpha values to transform Returns: list of percentiles corresponding to given alpha
-
expan.core.statistics.bootstrap(x, y, func=<function _delta_mean>, nruns=10000, percentiles=[2.5, 97.5], min_observations=20, return_bootstraps=False, relative=False)¶ Bootstraps the Confidence Intervals for a particular function comparing two samples. NaNs are ignored (discarded before calculation).
Parameters: - x (array like) – sample of treatment group
- y (array like) – sample of control group
- func (function) – function of which the distribution is to be computed. The default comparison metric is the difference of means. For bootstraping correlation: func=lambda x,y: np.stats.pearsonr(x,y)[0]
- nruns (integer) – number of bootstrap runs to perform
- percentiles (list) – The values corresponding to the given percentiles are returned. The default percentiles (2.5% and 97.5%) correspond to an alpha of 0.05.
- min_observations (integer) – minimum number of observations necessary
- return_bootstraps (boolean) – If this variable is set the bootstrap sets are returned otherwise the first return value is empty.
- relative (boolean) – if relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: - dict: percentile levels (index) and values
- np.array (nruns): array containing the bootstraping results per run
Return type: tuple
-
expan.core.statistics.chi_square(x, y, min_counts=5)¶ Performs the chi-square homogeneity test on categorical arrays x and y
Parameters: - x (array_like) – sample of the treatment variable to check
- y (array_like) – sample of the control variable to check
- min_counts (int) – drop categories where minimum number of observations or expected observations is below min_counts for x or y
Returns: - float: p-value
- float: chi-square value
- int: number of attributes used (after dropping)
Return type: tuple
-
expan.core.statistics.delta(x, y, assume_normal=True, percentiles=[2.5, 97.5], min_observations=20, nruns=10000, relative=False, x_weights=1, y_weights=1)¶ Calculates the difference of means between the samples (x-y) in a statistical sense, i.e. with confidence intervals.
NaNs are ignored: treated as if they weren’t included at all. This is done because at this level we cannot determine what a NaN means. In some cases, a NaN represents missing data that should be completely ignored, and in some cases it represents inapplicable (like PCII for non-ordering customers) - in which case the NaNs should be replaced by zeros at a higher level. Replacing with zeros, however, would be completely incorrect for return rates.
Computation is done in form of treatment minus control, i.e. x-y
Parameters: - x (array_like) – sample of a treatment group
- y (array_like) – sample of a control group
- assume_normal (boolean) – specifies whether normal distribution assumptions can be made
- percentiles (list) – list of percentile values for confidence bounds
- min_observations (integer) – minimum number of observations needed
- nruns (integer) – only used if assume normal is false
- relative (boolean) – if relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
- x_weights (list) – weights for the x vector, in order to calculate the weighted mean and confidence intervals, which is equivalent to the overall metric. This weighted approach is only relevant for ratios.
- y_weights (list) – weights for the y vector, in order to calculate the weighted mean and confidence intervals, which is equivalent to the overall metric. This weighted approach is only relevant for ratios.
Returns: - mu (float): mean value of the difference
- c_i (dict): percentile levels (index) and values
- ss_x (int): size of x excluding NA values
- ss_y (int): size of y excluding NA values
- _x (float): absolute mean of x
- _y (float): absolute mean of y
Return type: tuple
-
expan.core.statistics.estimate_std(x, mu, pctile)¶ Estimate the standard deviation from a given percentile, according to the z-score:
z = (x - mu) / sigmaParameters: - x (float) – cumulated density at the given percentile
- mu (float) – mean of the distribution
- pctile (float) – percentile value (between 0 and 100)
Returns: estimated standard deviation of the distribution
Return type: float
-
expan.core.statistics.normal_difference(mean1, std1, n1, mean2, std2, n2, percentiles=[2.5, 97.5], relative=False)¶ Calculates the difference distribution of two normal distributions.
Computation is done in form of treatment minus control. It is assumed that the standard deviations of both distributions do not differ too much.
Parameters: - mean1 (float) – mean value of the treatment distribution
- std1 (float) – standard deviation of the treatment distribution
- n1 (integer) – number of samples of the treatment distribution
- mean2 (float) – mean value of the control distribution
- std2 (float) – standard deviation of the control distribution
- n2 (integer) – number of samples of the control distribution
- percentiles (list) – list of percentile values to compute
- relative (boolean) – If relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: percentiles and corresponding values
Return type: dict
- For further information vistit:
- http://sphweb.bumc.bu.edu/otlt/MPH-Modules/BS/BS704_Confidence_Intervals/BS704_Confidence_Intervals5.html
-
expan.core.statistics.normal_percentiles(mean, std, n, percentiles=[2.5, 97.5], relative=False)¶ Calculate the percentile values for a normal distribution with parameters estimated from samples.
Parameters: - mean (float) – mean value of the distribution
- std (float) – standard deviation of the distribution
- n (integer) – number of samples
- percentiles (list) – list of percentile values to compute
- relative (boolean) – if relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: percentiles and corresponding values
Return type: dict
-
expan.core.statistics.normal_sample_difference(x, y, percentiles=[2.5, 97.5], relative=False)¶ Calculates the difference distribution of two normal distributions given by their samples.
Computation is done in form of treatment minus control. It is assumed that the standard deviations of both distributions do not differ too much.
Parameters: - x (array-like) – sample of a treatment group
- y (array-like) – sample of a control group
- percentiles (list) – list of percentile values to compute
- relative (boolean) – If relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: percentiles and corresponding values
Return type: dict
-
expan.core.statistics.normal_sample_percentiles(values, percentiles=[2.5, 97.5], relative=False)¶ Calculate the percentile values for a sample assumed to be normally distributed. If normality can not be assumed, use bootstrap_ci instead. NaNs are ignored (discarded before calculation).
Parameters: - values (array-like) – sample for which the normal distribution percentiles are computed.
- percentiles (list) – list of percentile values to compute
- relative (boolean) – if relative==True, then the values will be returned as distances below and above the mean, respectively, rather than the absolute values. In this case, the interval is mean-ret_val[0] to mean+ret_val[1]. This is more useful in many situations because it corresponds with the sem() and std() functions.
Returns: percentiles and corresponding values
Return type: dict
-
expan.core.statistics.pooled_std(std1, n1, std2, n2)¶ Returns the pooled estimate of standard deviation. Assumes that population variances are equal (std(v1)**2==std(v2)**2) - this assumption is checked for reasonableness and an exception is raised if this is strongly violated.
Parameters: - std1 (float) – standard deviation of first sample
- n1 (integer) – size of first sample
- std2 (float) – standard deviation of second sample
- n2 (integer) – size of second sample
Returns: Pooled standard deviation
Return type: float
- For further information visit:
- http://sphweb.bumc.bu.edu/otlt/MPH-Modules/BS/BS704_Confidence_Intervals/BS704_Confidence_Intervals5.html
- Todo:
- Also implement a version for unequal variances.
-
expan.core.statistics.sample_size(x)¶ Calculates sample size of a sample x :param x: sample to calculate sample size
Returns: sample size of the sample excluding nans Return type: int
expan.core.util module¶
-
expan.core.util.reindex(df, axis=0)¶ Parameters: - df –
- axis –
Returns:
Note
Partial fulfilment of: https://github.com/pydata/pandas/issues/2770
- Todo:
- test
- incorporate in pandas in drop() call and issue pull request
-
expan.core.util.scale_range(x, new_min=0.0, new_max=1.0, old_min=None, old_max=None, squash_outside_range=True, squash_inf=False)¶ Scales a sequence to fit within a new range.
If squash_inf is set, then infinite values will take on the extremes of the new range (as opposed to staying infinite).
- Args:
- x: new_min: new_max: old_min: old_max: squash_outside_range: squash_inf:
- Note:
- Infinity in the input is disregarded in the construction of the scale of the mapping.
>>> scale_range([1,3,5]) array([ 0. , 0.5, 1. ])
>>> scale_range([1,2,3,4,5]) array([ 0. , 0.25, 0.5 , 0.75, 1. ])
>>> scale_range([1,3,5, np.inf]) array([ 0. , 0.5, 1. , inf])
>>> scale_range([1,3,5, -np.inf]) array([ 0. , 0.5, 1. , -inf])
>>> scale_range([1,3,5, -np.inf], squash_inf=True) array([ 0. , 0.5, 1. , 0. ])
>>> scale_range([1,3,5, np.inf], squash_inf=True) array([ 0. , 0.5, 1. , 1. ])
>>> scale_range([1,3,5], new_min=0.5) array([ 0.5 , 0.75, 1. ])
>>> scale_range([1,3,5], old_min=1, old_max=4) array([ 0. , 0.66666667, 1. ])
>>> scale_range([5], old_max=4) array([ 1.])
expan.core.version module¶
-
expan.core.version.git_commit_count()¶ Returns the output of git rev-list –count HEAD as an int.
-
expan.core.version.git_latest_commit()¶ ” Returns output of git rev-parse HEAD.
-
expan.core.version.version(format_str='{short}')¶ Returns current version number in specified format.
Parameters: format_str (str) – Returns:
-
expan.core.version.version_numbers()¶